reduced dark and flat#
In order to perform the flat field correction (optional in nabu) an acquisition must contains reduced dark and flats.
Those reduced darks and flats comme from raw frames ‘darks’ and ‘flats’ frames. In general we expect those frames to be part of the NXtomo and call the dark and flat field construction widget to generate the reduced one.
reduced dark and flat field widget  #
#
This is usualy the first processing to run. This way the flat field correction can be done and is useful and / or required by many processes
By default the reduced dark(s) is obtained by computing the mean of raw dark frames and the reduced flat(s) is obtained by computing the median of the flat(s)
[1]:
from IPython.display import Video
Video("video/reduced_darks_flats_widget.mp4", embed=True, height=500)
[1]:
since tomoscan==1.0 (and nabu==2022.2, tomwer==1.0) reduced darks and flats are saved under
{dataset_prefix}_darks.hdf5and{dataset_prefix}_flats.hdf5files.for spec acquisition the reduced darks and flats will be duplicated (to dark.edf and refHSTXXXX.edf to insure backward compatibility
Here is a screenshot of a reduced_flats file containing a single serie of flat at the beginning
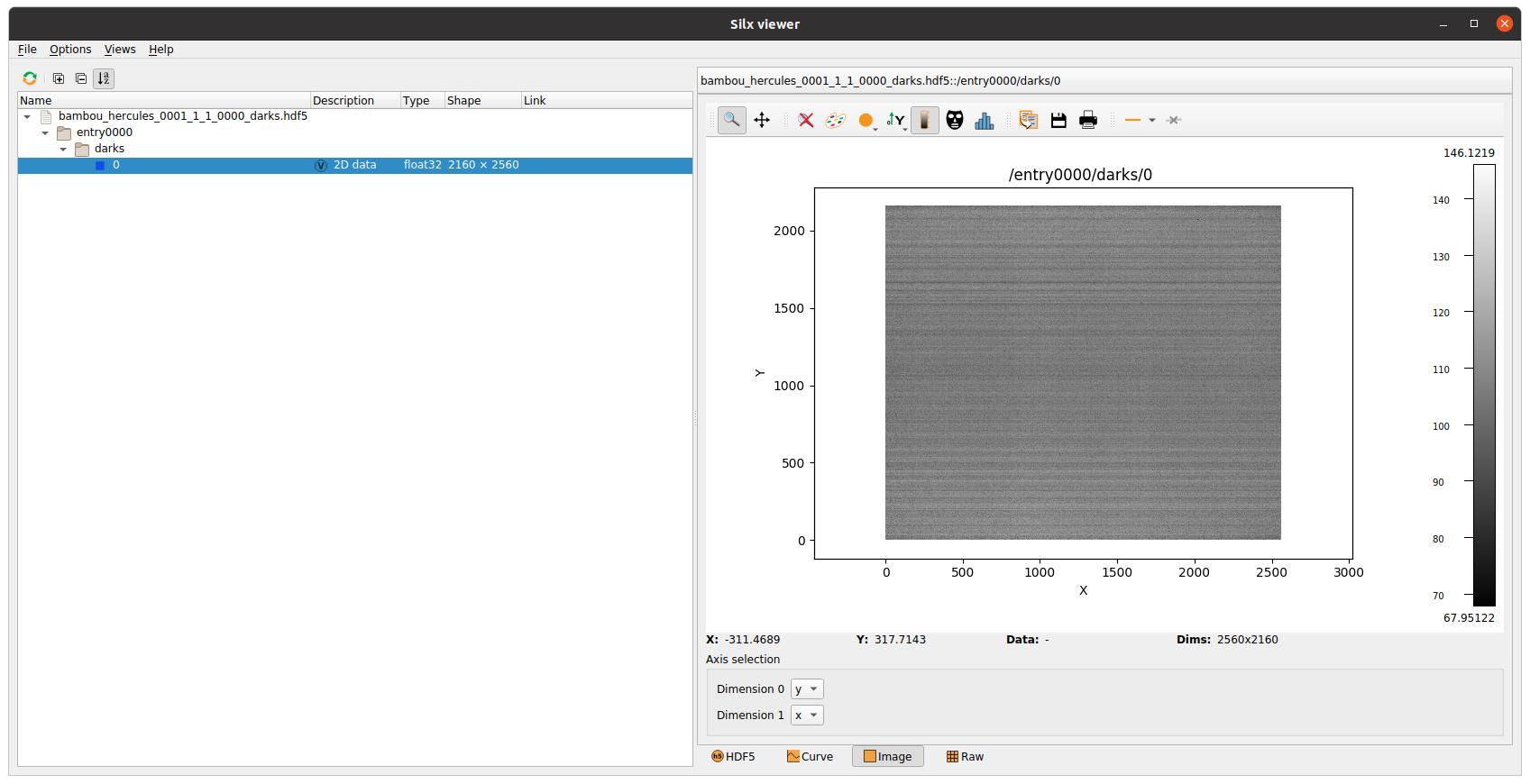
copying dark(s) and flat(s)#
In some cases it might happen that users need to reuse reduced darks or flats.
If the reduced darks and flats already exists then you can simply use the ‘copy’ from the reduced dark and flat widget
!!! The copy option is activated by default !!!
automode: Each time it mets a dataset with reduced dark / flat it will keep them in cache. And when it mets a dataset with dark / flat missing it will copy them to it.
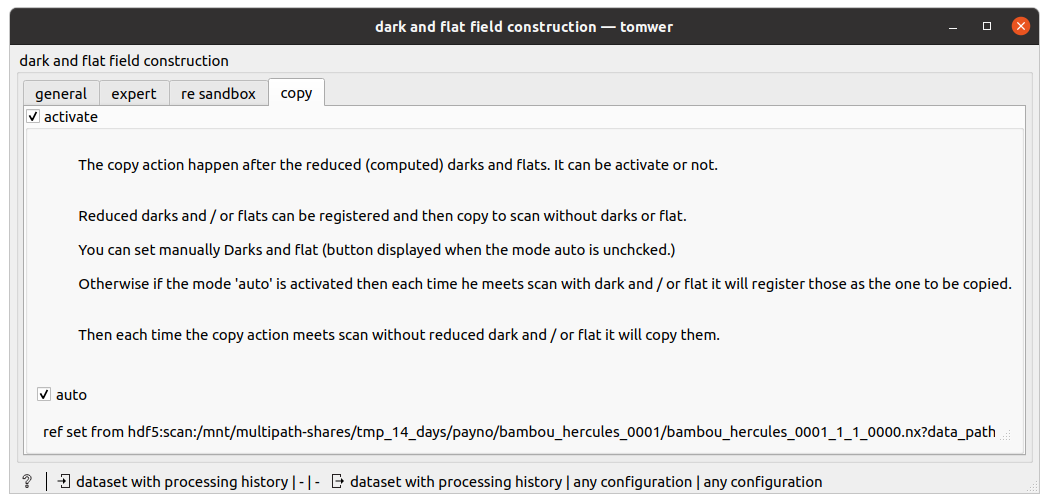
manualmode: user can provide an URL to well formed reduced darks and reduced flats HDF5 dataset
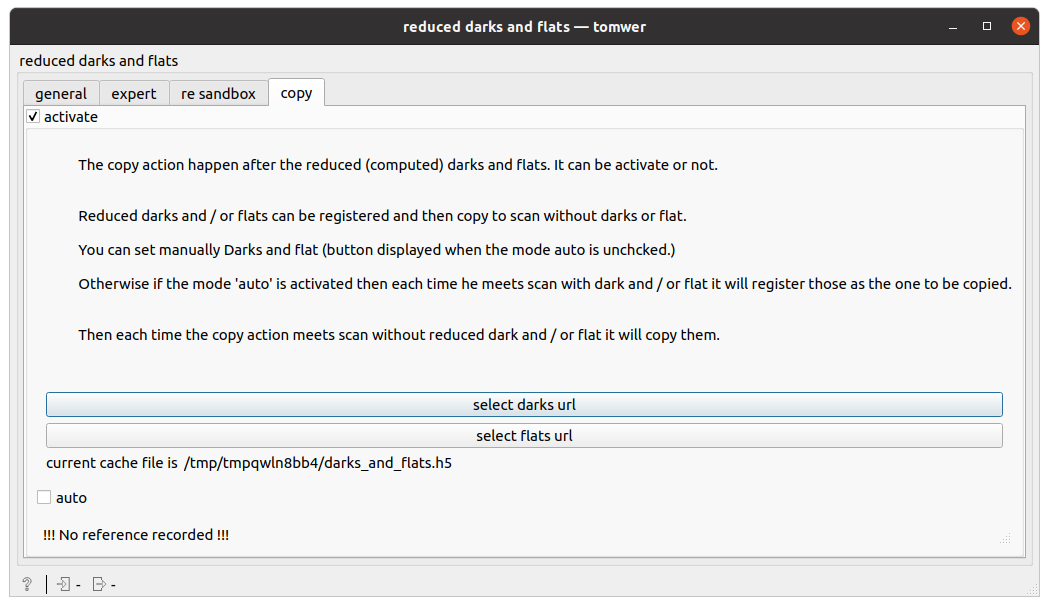
note: the darks and flats cache file is provided at the bottom of the widget. This can be a good to check the registration goes as expected.
from a python script#
You can also create those following the tomoscan python API like
provide reduced darks and flats as a dictionary (key is the index and value is a 2D numpy array)
from tomwer.core.scan.nxtomoscan import NXtomoScan
# from tomwer.core.scan.edfscan import EDFTomoScan
# same API for EDF or HDF5
scan = NXtomoScan(file_path, data_path)
darks = {
0: numpy.array(...), # darks at start
}
flats = {
0: numpy.array(...), # flats at start
3000: numpy.array(...), # flats at end
}
scan.save_reduced_darks(darks)
scan.save_reduced_flats(flats)
provide reduced darks and flats from already existing HDF5 dataset
from tomoscan.esrf.scan.utils import copy_darks_to, copy_flats_to
# create darks and flats as numpy array
darks = {
0: numpy.ones((100, 100), dtype=numpy.float32),
}
flats = {
1: numpy.ones((100, 100), dtype=numpy.float32) * 2.0,
100: numpy.ones((100, 100), dtype=numpy.float32) * 2.0,
}
original_dark_flat_file = os.path.join(tmp_path, "originals.hdf5")
dicttoh5(darks, h5file=original_dark_flat_file, h5path="darks", mode="a")
dicttoh5(flats, h5file=original_dark_flat_file, h5path="flats", mode="a")
# create darks and flats URL
darks_url = DataUrl(
file_path=original_dark_flat_file,
data_path="/darks",
scheme="silx",
)
flats_url = DataUrl(
file_path=original_dark_flat_file,
data_path="/flats",
scheme="silx",
)
# apply the copy
scan = NXtomoScan(...)
copy_flats_to(scan=scan, flats_url=flats_url, save=True)
copy_flats_to(scan=scan, flats_url=flats_url, save=True)
computing reduced flats from projection#
It can happen that sometime you have a bliss dataset containing projections that must be used as flats in order to compute the reduced flats (dataset 1). And that this reduced flats must be used by other datasets (datasets 2*).
In this case you can do the following actions:
preprocessing (computing reduced flats from dataset 1)
convert dataset 1 NXtomo image_key projections to flats (using
image-key-editororimage-key-upgraderwidget.then compute reduced flat from those raw flats (using
reduced dark and flatwidget ).copy this reduced flats to the datasets 2 (by providing url to the reduced flats or setting them directly from the
reduced flatsinput as show in the video)
processing (reconstructing datasets 2*)
compute reduced darks (from raw) and copy reduced flats from dataset 1 using the
reduced dark and flatwidget (reduced flathave been set during pre processing.then create the workflow you want to process (
default center of rotation,nabu sliceanddata vieweron the video)
[2]:
from IPython.display import YouTubeVideo
YouTubeVideo("vJOo0rHHUYk", height=500, width=800)
[2]:
Note: if the processing is done before the flat copy is done or if this one fails then flat field will fail. And you might encounter the following error:
2023-03-06 16:04:45,967 [ERROR] cannot make flat field correction, flat not found [tomwer.core.scan.scanbase](scanbase.py:177)
2023-03-06 16:04:45,967:ERROR:tomwer.core.scan.scanbase: cannot make flat field correction, flat not found
hand on - exercise A#
The /scisoft/tomo_training/part3_flatfield/WGN_01_0000_P_110_8128_D_129/ contains three NXtomo.
Use the first one (WGN_01_0000_P_110_8128_D_129_0000.nx) projections as flats to compute reduced flats.
Then provide this reduced flat to compute one of the two other dataset (WGN_01_0000_P_110_8128_D_129_0001.nx or WGN_01_0000_P_110_8128_D_129_0002.nx)
Note: this dataset is provided as a proof of concept. Please don’t be very ‘attentive’ to the slice reconstruction
