tomwer z-stitching application#
Step 0: connect to a slurm node (if you want to run stitching remotly) and activate a python environment#
connect to a node#
For this use case the usage of the ‘interactive’ partition is recommended. Here we are using the GUI so you will also need X forwarding.
Depending on the dimensions of the camera and the number of scans / volumes you might need more or less memory.
ssh -XC cluster-access
salloc --partition interactive --mem=256G --x11 srun --pty bash -l
activate a python environment containing tomotools#
For example we can use the ‘stable’ version of the tomotools.
module load tomotools/stable
Then you should have access to all tomotools software. You can check it by doing
which nabu
which nxtomomill
which tomwer
those commands should return the path of the applications.
Step 1: launch tomwer z-stitching with a set of NXtomos or Volumes to be stitched#
In this example we want to stitch together a set of NXtomos together (from a z-serie). We consider that we already have all the NXtomo files under PROCESSED_DATA/{COLLECTION}/{DATASET}/*.nx
if you don’t have the NXtomos then you can have a look at nxtomomill batch processing which present you how you can batch call nxtomomill h52nx
1.1 launch the application#
So to launch it you can simply go for
[1]:
# ! tomwer z-stitching PROCESSED_DATA/{COLLECTION}/{DATASET}/*.nx
you can access help of the application by using nxtomomill h52nx --help
It will load metadata of all requested scan (or volume). And will open a window with all this information
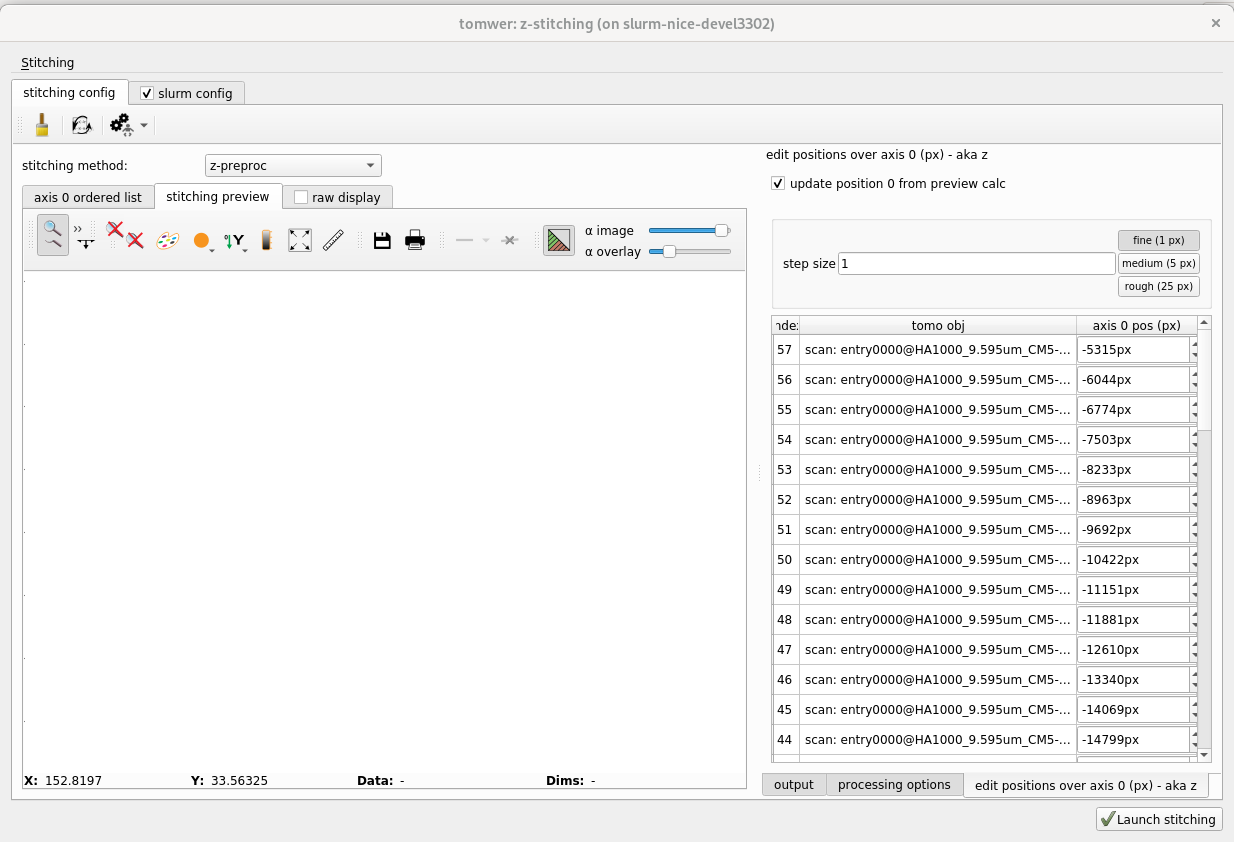
On the right all the scans or volumes must be there. If there is some missing you can add them from stiching -> add tomo object
1.2 set up the stitching#
Once you have all objects that you want to stitch:
1.2.1 check that the axis positions has been found and make sense. Else edit them.#
the axis positions are available from the panel on the right edit positions over axis 0 They should obtained automatically from the scan or volume metadata. (volume must have been reconstructed with nabu 2024 or higher) If this is not the case you can still provide them manually.
1.2.2 tune the stitching settings.#
Once the position seems (approximatively) correct you can define tune the stitching to be done. You can access it from the panel on the right ‘processing options’ Here we will only present the ‘basic’ parameters only
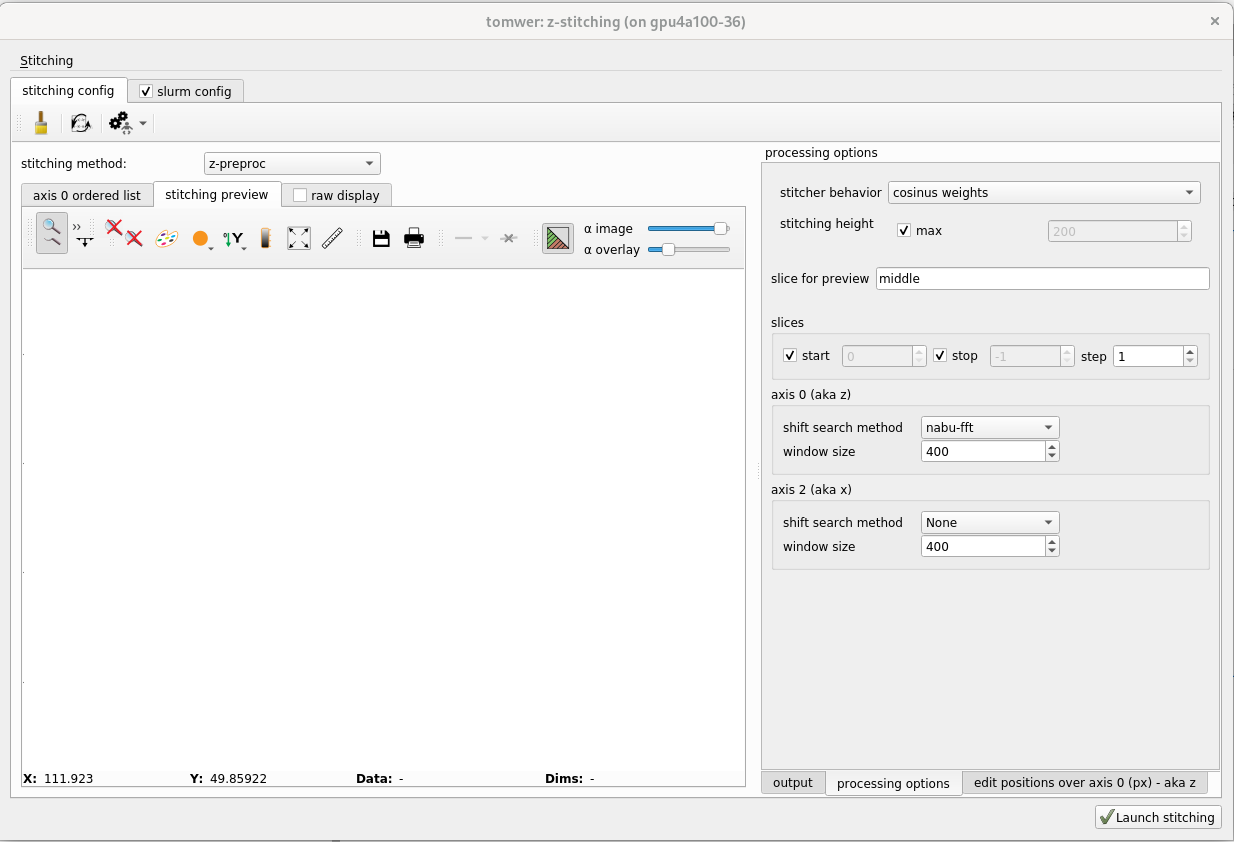
For example by default the overlap will be created on the largest region possible. But sometime it can be interesting to reduce the size to have a sharper image.
To modify it uncheck ‘stitching height / max’ and provide the value.
motor position refinement#
The motor position can be refine by ‘shift search’ algorithm. For each overlap it will pick the two images at overlap region and will call one of the cross-correlation algorithm (shift-search method).
If no None algorithm is defined then there will be no refinement.
1.2.3 process some preview#
Once you are happy with the settings you can ask to create a preview of the stitching.
This will call the exact same code as the stitching of the entire volume. And so you will be able to determine if you are happy with the parameters or not.
For it you can either press the dedicated button: or by the ‘F5’ shortcut
Then tune stitching configuration, scans / volumes positions until you are happy with the preview.

the stiching of scans will be done within flat field correction. So you need to deactivate it when you call nabu
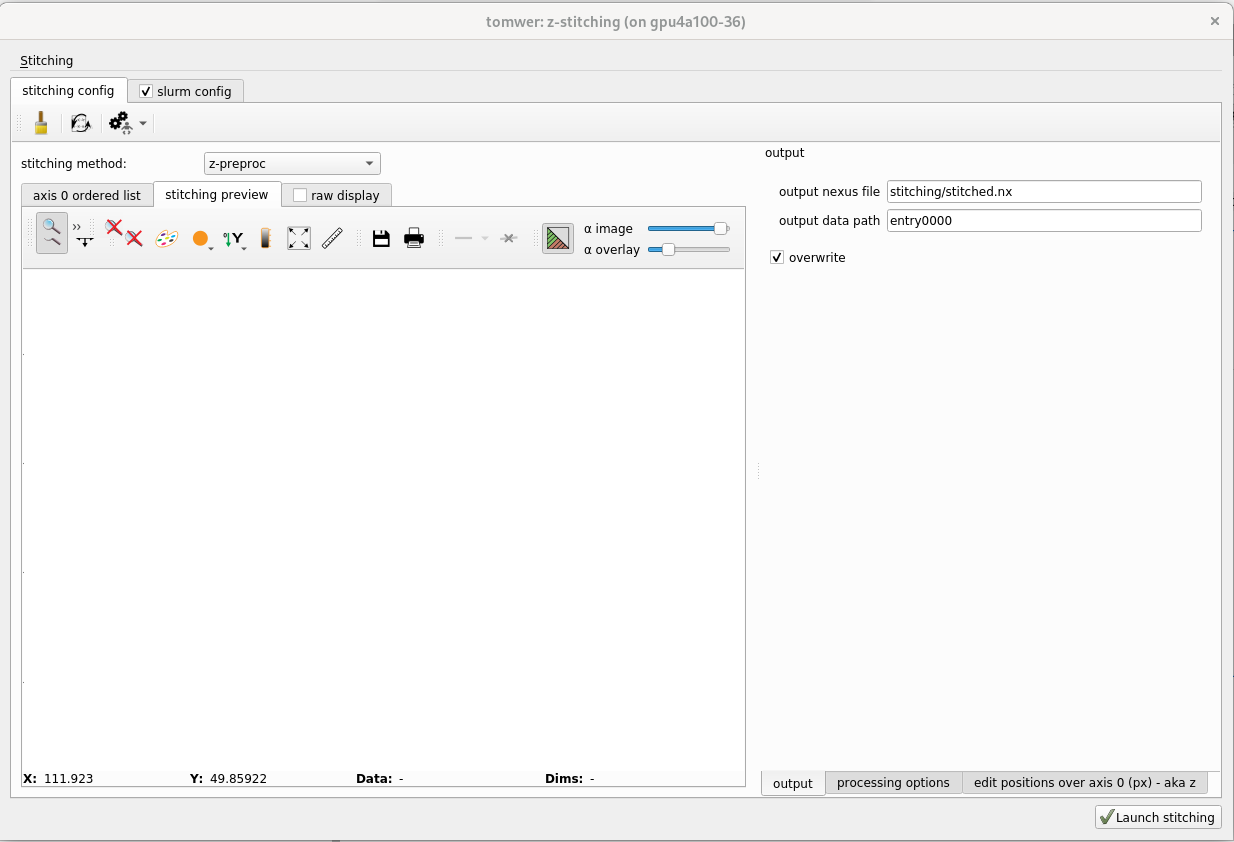
1.2.4 define the stiching out#
When you are happy with the settings and the preview it is time to launch the stitching of the full volumes / set of projections.
Define the output of the stitching from the panel on the right ‘output’
2. run the stiching#
2.1 (optional) slurm configuration#
if the computer executing the application is a slurm client then you can submit the stitching to slurm. In this case the ‘slurm config’ tab will be unable and you can check it. It will activate stiching submission to slurm.
GUI slurm config disabled |
GUI slurm config enabled |
|---|---|
|
|
Then from this interface you can tune the stitching job to be done.
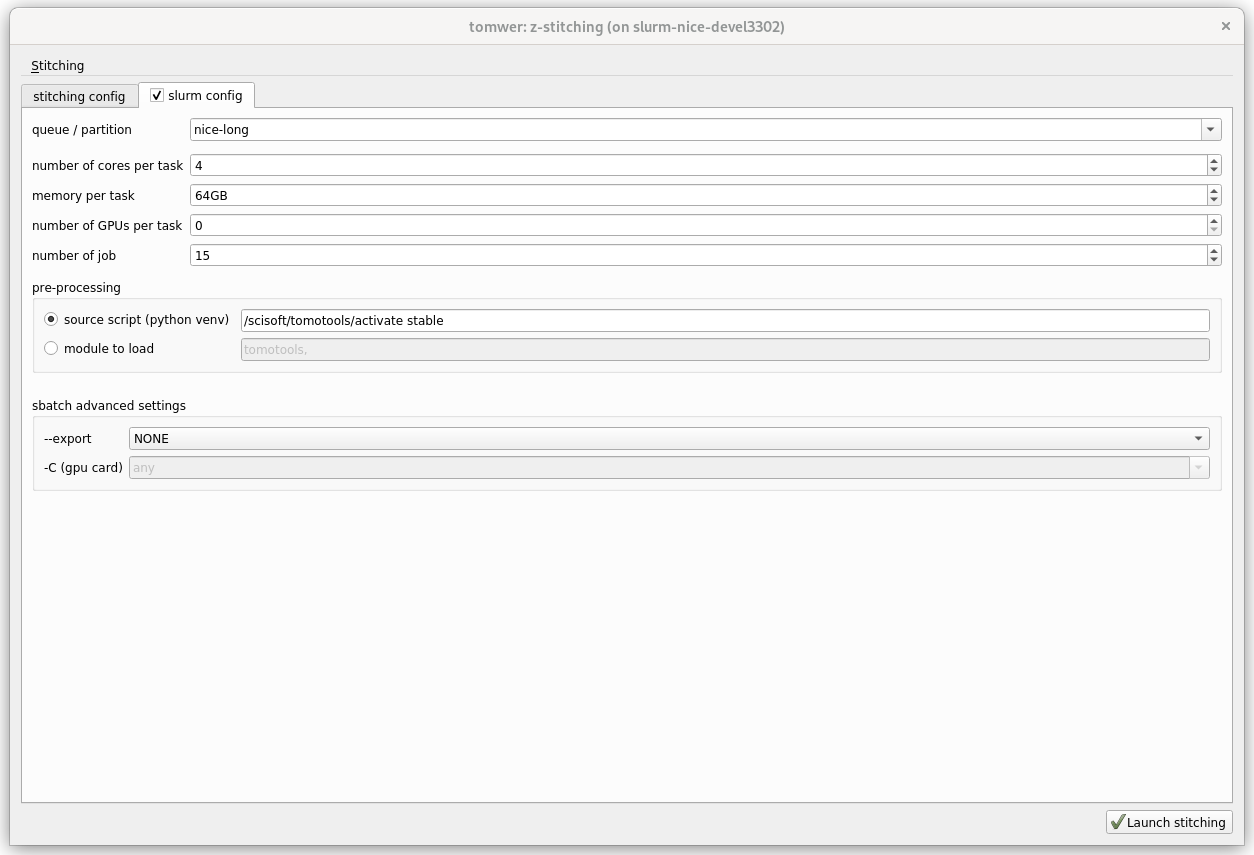
queue#
one which slurm partition the jobs will be submitted
number of cores per task#
number of cpu core for each job
memory per task#
memory of each job. Depending on the size of the slices you might need to increase this size.
number of GPUs per task#
Stitching only works on CPU so please always set it to 0
number of job#
How much job should we create to stitch the projections of the volume.
Each job will stitch along the axis 2 (z) a part of the slices. Projections stitching will be divided along projections, volume stitching along axis 1 (aka y) Then when all jobs are finished a ‘master file’ will be created (in the case of HDF5) to connect them all together.
Example 1: we have 3 set of 2kx2k projections of 1000 angles and we ask for 2 jobs then each job will handle 500 projections
Example 2: we want to stitch two 1kx2kx3k volumes (z == axis 0 == 1k) on 4 jobs. Then each job will handle 500 slices.
pre-processing#
When you submit a job to slurm you must provide a way to get a python environment knowing the python libraries (nabu…). Then for this you can either source a script or load some modules. Usually the simpler is to use the same method as you access it to launch the application. It will also ensure safe compatibility.
2.2 launch stitching#
Press ‘Launch stitching’.
in the case of distributed stitching (slurm)#
It will split the stitching into number of job. Then wait until all jobs are finished.
Then if all jobs are finished and suceed it will create a master file to link all the job sub volume / sub NXtomo togther.
Distributed stitching will create a sub folder (stitching/stitched by default) with each job specific configuration, script submitted (slurm job) and output created.
It might be possible that you will be quick out before all job are runned. But those job will be continue. If it happens you can run the sub parts aggregation in post processing by calling nabu-stitching [configuration-file] --only-create-master-file.
If some job failed for any reason you can also relaunch them manually and independently. Then launch the post processing aggregation (see upper)
in the case of local stitching#
Stitching will be processing into a single file.
3. check the result#
You can observer the result using a tool like silx view [stitched.nx | stitched.hdf5] …
Please if you see any issue / encounter any scory please let us know
( 4. process reconstruction - in the case of stiching of scans / over projections)#
If your stitching was done on projections you will end up with an NXtomo fully compatible with tomotools and you can reconstruct it using nabu / tomwer
[2]:
# ! nabu-config --dataset [stitched.nx] --output nabu_recons_config.conf
# ! nabu nabu_recons_config
And of course you can browse your recoonstruction with a tool like silx or imageJ or …
extra#
Some videos of an earlier prototype are available from tomotools’s stiching yoututbe playlist