Publish processed data to icat#
publishing from tomwer#
Since tomwer 1.3 some task are embeded to publish processed data to icat. Associated widgets / tasks are:
An example of workflow using the three widgets is available in help -> example -> ‘Reconstruction of a volume and publication to icat’
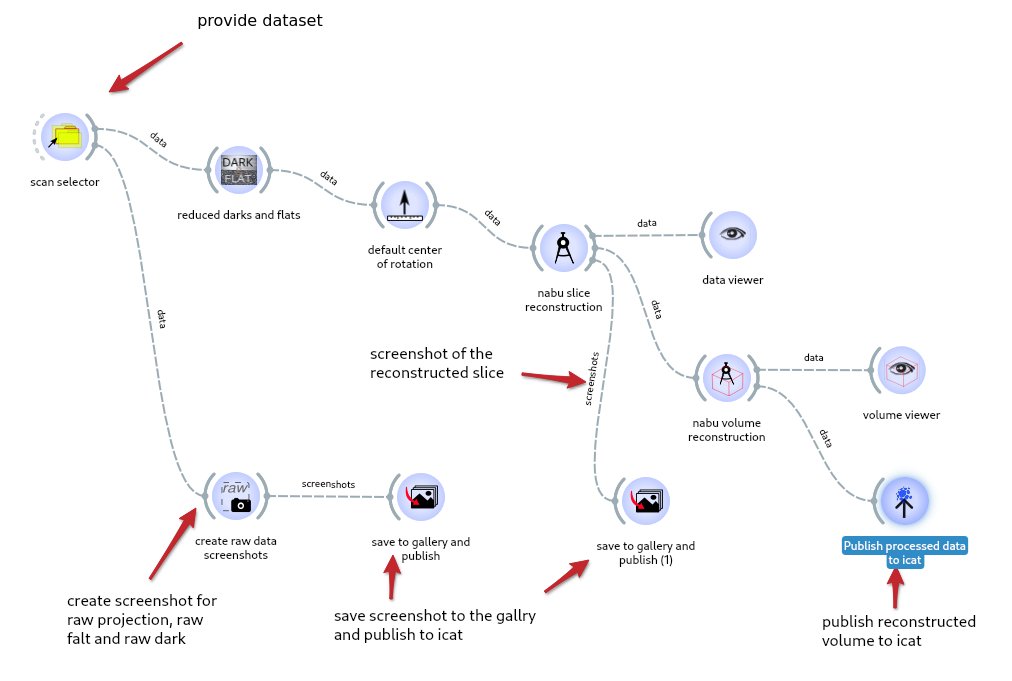
example of a workflow publishing the gallery and the processed volume#
The input dataset must be under a “PROCESSED_DATA” folder else publication will not be done.
Only NXtomo input are considered for now (no EDF)
All necessary metadata should be retrieve from the .nx file (dataset, beamline…) if this fail user can style provide them …
Note
For publishing to icat and have processing clearly decouple we also added a folder ‘volume reconstruction’ for volume recosntruction
publishing from CLI#
The publication to icat can also be done from the Command Line Interface (CLI) using the pyicat-plus project
The software is embed by tomwer so you can access it with the same python environment.
To store processed data you can go for something like:
icat-store-processed --beamline id00 \
--proposal id002207 \
--path /data/visitor/path/to/processed/data \
--dataset testproc \
--sample mysample \
--raw /data/visitor/path/to/dataset1
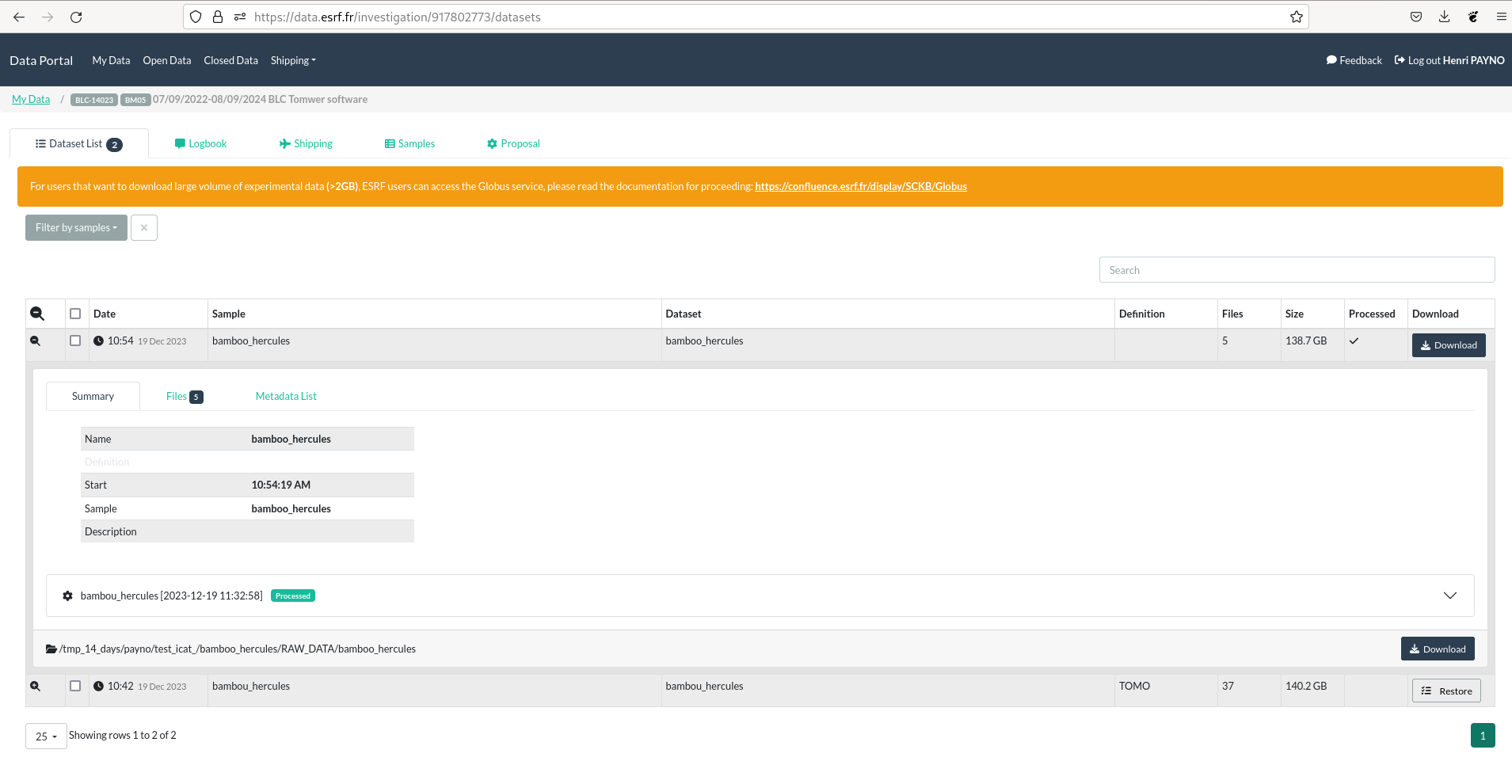
data.esrf.fr#
Once your dataset have been publish you should be able to retrieve them from https://data.esrf.fr/
The differente processed dataset should be linked to the raw data.
The raw data publication is handled automatically by bliss